Professional Manufacturer of Biomagnetic Beads

How doLarge DNA Fragment Selectiontechnologyin NGS Sequencing
Large DNA fragment selection technology plays a critical role in NGS sequencing, particularly in structural variant detection, genome assembly, and haplotype analysis. Below are its core applications and technical principles::
I. Application Scenarios
Structural Variant (SV) Detection
Challenge: Conventional short-read sequencing (e.g., Illumina) struggles to accurately capture variants >1 kb (insertions, deletions, inversions, translocations).
Solution: By selecting large DNA fragments (10–50 kb) for library construction, combined with long-read sequencing (PacBio/Nanopore) or linked-read analysis, complex SVs spanning repetitive regions can be resolved.
Example: Fusion events in cancer genomes (e.g., *ALK-EML4*) often require large-fragment data.
High-Quality Genome Assembly
Challenge: Short-read data often leads to assembly errors in repetitive regions, causing genome fragmentation.
Solution: Large fragments (e.g., Fosmid libraries) provide long-range physical linkage, enabling continuous scaffold assembly (5–10× improvement in N50).
Example: The T2T Consortium filled the final 8% gap in the human reference genome using Hi-C and ultra-long-read technologies.
Haplotype Phasing
Challenge: Short-read sequencing fails to preserve allelic linkage at heterozygous sites.
Solution: Barcode-based technologies (e.g., 10X Genomics) reconstruct haplotype blocks spanning hundreds of kb.
Value: Critical for HLA typing and identifying disease-causing gene linkages.
II. Core Selection Technologies
1. Physical Separation Methods
Pulse Field Gel Electrophoresis (PFGE): Separates fragments >50 kb for metagenomic or complex sample screening.
Magnetic Bead Selection (SPRIselect): Recovers specific fragment sizes (e.g., 0.1–10 kb or >15 kb) by adjusting bead-to-sample ratios.
2. Barcode-Based Enrichment
10X Genomics Chromium: Encapsulates DNA fragments in oil droplets, labeling short reads from the same large fragment with barcodes.
TELL-Seq/Linked-Reads: Uses microfluidics for virtual long-fragment reconstruction at 50% lower cost than 10X.
3. Circularization-Based Amplification
Loop Genomics: DNA fragments are circularized and amplified via rolling circle replication, preserving physical adjacency for standard NGS platforms.
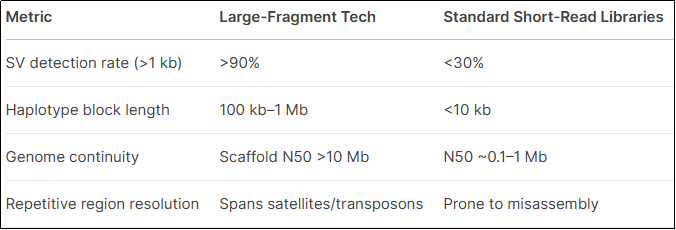
III. Advantages vs. Conventional Methods
IV. Challenges and Optimization
High DNA Quality Required: Needs intact DNA (DV200 >50%), incompatible with FFPE samples to Development of low-damage extraction kits.
Cost Control: Long-read sequencing remains expensive to Hybrid strategies (e.g., Linked-Reads and Illumina) reduce costs.
Bioinformatics Upgrades: Specialized tools needed (e.g., Canu for assembly, HapCUT2 for phasing).
V. Representative Applications
COVID-19 Tracing: Nanopore sequencing and magnetic bead selection (>20 kb fragments) rapidly resolved SARS-CoV-2 genomes and recombination events.
Genetic Disease Diagnosis: 10X Genomics detected SMN1 exon 7 deletions, avoiding interference from SMN2.
Plant Genomics: In barley genome assembly, BAC libraries (40–100 kb) increased Contig N50 to 486 kb.
Conclusion
Large DNA fragment selection overcomes the limitations of short-read NGS by providing long-range genomic context. With advances in barcoding (e.g., TELL-Seq) and localized platforms (e.g., MGI CoolMPS), this technology will accelerate applications in clinical diagnostics, evolutionary studies, and precision breeding.
Supplier
Shanghai Lingjun Biotechnology Co., Ltd. was established in 2016 which is a professional manufacturer of biomagnetic materials and nucleic acid extraction reagents.
We have rich experience in nucleic acid extraction and purification, protein purification, cell separation, chemiluminescence, and other technical fields.
Our products are widely used in many fields, such as medical testing, genetic testing, university research, genetic breeding, and so on. We not only provide products but also can undertake OEM, ODM, and other needs. If you have a related need, please feel free to contact us .